I would like to train a GCN for protien-ligand binding affinity regression. I use GCNConv from pytorch geometric, ReLU for all activations and Dropout (0.2) after 2 dense layers each. Using ReLU for all activation functions.
While my training curves look overall at the same level  , i noticed weird periodic jumps at validation stage
, i noticed weird periodic jumps at validation stage 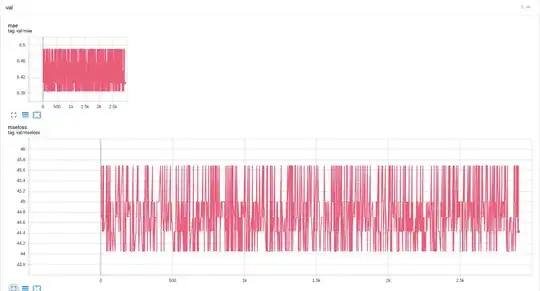 . What might be the issue with the model or/and training process?
. What might be the issue with the model or/and training process?
For me it seems that the network starts initializing again after each epoch, but it is not what my code seems like doing.