Yes, through the components_ property:
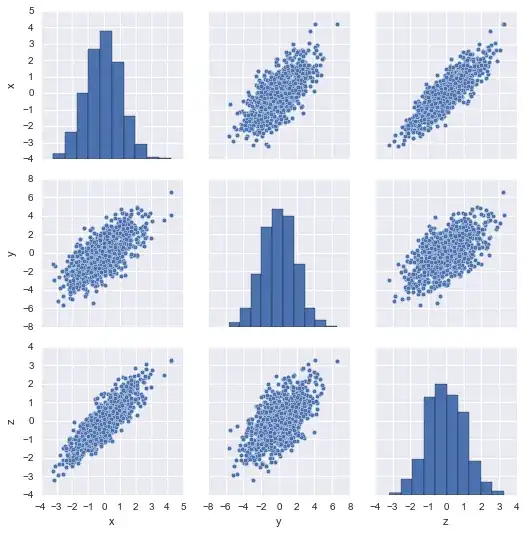
import numpy, seaborn, pandas, sklearn.decomposition
data = numpy.random.randn(1000, 3) @ numpy.random.randn(3,3)
seaborn.pairplot(pandas.DataFrame(data, columns=['x', 'y', 'z']));
sklearn.decomposition.RandomizedPCA().fit(data).components_
> array([[ 0.43929754, 0.81097276, 0.38644644],
[-0.54977152, 0.58291122, -0.59830243],
[ 0.71047094, -0.05037554, -0.70192119]])
sklearn.decomposition.RandomizedPCA(2).fit(data).components_
> array([[ 0.43929754, 0.81097276, 0.38644644],
[-0.54977152, 0.58291122, -0.59830243]])
We see that the truncated decomposition is simply the truncation of the full decomposition. Each row contains the coefficients of the corresponding principal component.
