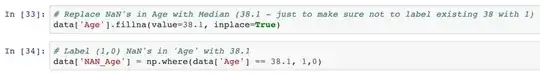
To decide which strategy is appropriate, it is important to investigate the mechanism that led to the missing values to find out whether the missing data is missing completely at random (MCAR), missing at random (MAR), or missing not at random (MNAR).
- MCAR means that there is no relationship between the missingness of the data and any of the values.
- MAR means that that there is a systematic relationship between the propensity of missing values and the observed data, but not the missing data.
- MNAR means that there is a systematic relationship between the propensity of a value to be missing and its values.
Given what you have told its likely that its MCAR. (assumption is that you already tried to find this propensity yourself (domain knowledge) or build a model between the missing columns and other features and failed in doing so)
Some other techniques to impute the data, I would suggest looking at KNN imputation (from experience always solid results) but you should try different methods
fancy impute supports such kind of imputation, using the following API:
from fancyimpute import KNN
# Use 10 nearest rows which have a feature to fill in each row's missing features
X_fill_knn = KNN(k=10).fit_transform(X)
Here are different methods also supported by this package:
•SimpleFill: Replaces missing entries with the mean or median of each
column.
•KNN: Nearest neighbor imputations which weights samples using the
mean squared difference on features for which two rows both have
observed data.
•SoftImpute: Matrix completion by iterative soft thresholding of SVD
decompositions. Inspired by the softImpute package for R, which is
based on Spectral Regularization Algorithms for Learning Large
Incomplete Matrices by Mazumder et. al.
•IterativeSVD: Matrix completion by iterative low-rank SVD
decomposition. Should be similar to SVDimpute from Missing value
estimation methods for DNA microarrays by Troyanskaya et. al.
•MICE: Reimplementation of Multiple Imputation by Chained Equations.
•MatrixFactorization: Direct factorization of the incomplete matrix
into low-rank U and V, with an L1 sparsity penalty on the elements of
U and an L2 penalty on the elements of V. Solved by gradient descent.
•NuclearNormMinimization: Simple implementation of Exact Matrix
Completion via Convex Optimization by Emmanuel Candes and Benjamin
Recht using cvxpy. Too slow for large matrices.
•BiScaler: Iterative estimation of row/column means and standard
deviations to get doubly normalized matrix. Not guaranteed to converge
but works well in practice. Taken from Matrix Completion and Low-Rank
SVD via Fast Alternating Least Squares.
EDIT: MICE was deprecated and they moved it to sklearn under iterative imputer